| Synonym | CD66c, CEACAM6, Normal cross-reacting antigen, NCA, CEAL, Nonspecific crossreacting antigen, CD66c antigen |
|---|
| Refseq ID | NM_002483 |
|---|
| GeneID | 4680 |
|---|
| Unigene ID | Hs.466814 |
|---|
| Probe set ID | 203757_s_at |
|---|
|
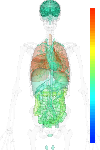 |
| EST |
|
brain:- | |
blood:75.60 | |
connective:54.60 | |
reproductive:13.70 | |
muscular:- | |
alimentary:503.00 | |
liver:- | |
lung:97.80 | |
urinary:- | |
endo/exo-crine:104.80 | |
|
| GeneChip |
|
brain:4.07 | |
blood:5.01 | |
connective:4.47 | |
reproductive:4.42 | |
muscular:4.23 | |
alimentary:6.10 | |
liver:4.17 | |
lung:9.40 | |
urinary:3.86 | |
endo/exo-crine:5.22 | |
|
| CAGE |
|
brain:0.02 | |
blood:1.69 | |
connective:0.00 | |
reproductive:1.06 | |
muscular:0.65 | |
alimentary:3.89 | |
liver:1.12 | |
lung:5.66 | |
urinary:1.05 | |
endo/exo-crine:0.93 | |
|
| RNA-seq |
|
brain:0.02 | |
blood:0.60 | |
connective:1.76 | |
reproductive:0.61 | |
muscular:0.00 | |
alimentary:1.51 | |
liver:0.23 | |
lung:7.24 | |
urinary:0.04 | |
endo/exo-crine:0.15 | |
|
| Synonym | SIR2L, SIR2-like protein 2, NAD-dependent deacetylase sirtuin-2, SIRT2, SIR2-like, SIR2L2 |
|---|
| Refseq ID | NM_012237 |
|---|
| GeneID | 22933 |
|---|
| Unigene ID | Hs.466693 |
|---|
| Probe set ID | 220605_s_at |
|---|
|
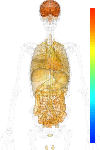 |
| EST |
|
brain:364.70 | |
blood:80.80 | |
connective:125.30 | |
reproductive:99.90 | |
muscular:367.00 | |
alimentary:80.50 | |
liver:21.10 | |
lung:48.90 | |
urinary:115.20 | |
endo/exo-crine:62.10 | |
|
| GeneChip |
|
brain:9.09 | |
blood:7.94 | |
connective:8.08 | |
reproductive:7.78 | |
muscular:8.87 | |
alimentary:8.30 | |
liver:8.17 | |
lung:8.06 | |
urinary:8.09 | |
endo/exo-crine:8.03 | |
|
| CAGE |
|
brain:5.13 | |
blood:3.45 | |
connective:3.95 | |
reproductive:3.55 | |
muscular:4.34 | |
alimentary:3.68 | |
liver:4.23 | |
lung:3.56 | |
urinary:3.61 | |
endo/exo-crine:3.84 | |
|
| RNA-seq |
|
brain:1.90 | |
blood:0.00 | |
connective:0.22 | |
reproductive:1.79 | |
muscular:2.61 | |
alimentary:1.71 | |
liver:0.54 | |
lung:0.52 | |
urinary:1.75 | |
endo/exo-crine:1.57 | |
|
| Synonym | SIR2L, SIR2-like protein 2, NAD-dependent deacetylase sirtuin-2, SIRT2, SIR2-like, SIR2L2 |
|---|
| Refseq ID | NM_030593 |
|---|
| GeneID | 22933 |
|---|
| Unigene ID | Hs.466693 |
|---|
| Probe set ID | 220605_s_at |
|---|
|
 |
| EST |
|
brain:364.70 | |
blood:80.80 | |
connective:125.30 | |
reproductive:99.90 | |
muscular:367.00 | |
alimentary:80.50 | |
liver:21.10 | |
lung:48.90 | |
urinary:115.20 | |
endo/exo-crine:62.10 | |
|
| GeneChip |
|
brain:9.09 | |
blood:7.94 | |
connective:8.08 | |
reproductive:7.78 | |
muscular:8.87 | |
alimentary:8.30 | |
liver:8.17 | |
lung:8.06 | |
urinary:8.09 | |
endo/exo-crine:8.03 | |
|
| CAGE |
|
brain:5.13 | |
blood:3.45 | |
connective:3.95 | |
reproductive:3.55 | |
muscular:4.34 | |
alimentary:3.68 | |
liver:4.23 | |
lung:3.56 | |
urinary:3.61 | |
endo/exo-crine:3.84 | |
|
| RNA-seq |
|
brain:5.32 | |
blood:3.03 | |
connective:3.92 | |
reproductive:3.77 | |
muscular:4.03 | |
alimentary:3.52 | |
liver:2.48 | |
lung:3.54 | |
urinary:2.97 | |
endo/exo-crine:4.02 | |
|
| Synonym | 26S proteasome regulatory subunit S14, Nin1p, HIP6, S14, PSMD8, HYPF, MGC1660, 26S proteasome non-ATPase regulatory subunit 8, p31 |
|---|
| Refseq ID | NM_002812 |
|---|
| GeneID | 5714 |
|---|
| Unigene ID | - |
|---|
| Probe set ID | 200820_at |
|---|
|
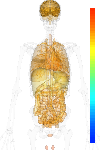 |
| EST |
nodata |
|
| GeneChip |
|
brain:8.17 | |
blood:8.15 | |
connective:8.90 | |
reproductive:8.67 | |
muscular:8.67 | |
alimentary:8.32 | |
liver:7.69 | |
lung:8.26 | |
urinary:8.22 | |
endo/exo-crine:8.28 | |
|
| CAGE |
|
brain:4.96 | |
blood:4.93 | |
connective:5.38 | |
reproductive:5.06 | |
muscular:5.68 | |
alimentary:5.04 | |
liver:5.60 | |
lung:4.86 | |
urinary:5.02 | |
endo/exo-crine:5.02 | |
|
| RNA-seq |
|
brain:4.83 | |
blood:5.13 | |
connective:5.94 | |
reproductive:5.43 | |
muscular:6.50 | |
alimentary:4.90 | |
liver:5.54 | |
lung:5.47 | |
urinary:5.53 | |
endo/exo-crine:5.48 | |
|
| Synonym | NADPH-flavin reductase, MGC117413, GHBP, BVR-B, Biliverdin reductase B, NADPH-dependent diaphorase, Green heme-binding protein, Biliverdin-IX beta-reductase, BVRB, BLVRB, biliverdin reductase B (flavin reductase (NADPH)), Flavin reductase, FLR, FR |
|---|
| Refseq ID | NM_000713 |
|---|
| GeneID | 645 |
|---|
| Unigene ID | Hs.515785 |
|---|
| Probe set ID | 202201_at |
|---|
|
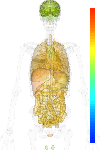 |
| EST |
|
brain:32.10 | |
blood:36.50 | |
connective:41.80 | |
reproductive:24.60 | |
muscular:124.90 | |
alimentary:30.20 | |
liver:116.10 | |
lung:68.40 | |
urinary:- | |
endo/exo-crine:101.00 | |
|
| GeneChip |
|
brain:7.04 | |
blood:7.97 | |
connective:8.43 | |
reproductive:7.64 | |
muscular:8.29 | |
alimentary:8.19 | |
liver:8.85 | |
lung:7.98 | |
urinary:8.08 | |
endo/exo-crine:7.71 | |
|
| CAGE |
|
brain:3.12 | |
blood:4.09 | |
connective:4.66 | |
reproductive:3.81 | |
muscular:4.28 | |
alimentary:4.37 | |
liver:5.13 | |
lung:4.52 | |
urinary:4.44 | |
endo/exo-crine:3.66 | |
|
| RNA-seq |
|
brain:3.56 | |
blood:5.11 | |
connective:5.49 | |
reproductive:5.10 | |
muscular:5.44 | |
alimentary:5.31 | |
liver:6.41 | |
lung:6.00 | |
urinary:5.12 | |
endo/exo-crine:5.51 | |
|
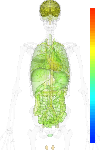
| Synonym | LIPE, lipase, hormone-sensitive, LHS, Hormone-sensitive lipase, HSL |
|---|
| Refseq ID | NM_005357 |
|---|
| GeneID | 3991 |
|---|
| Unigene ID | - |
|---|
| Probe set ID | 208186_s_at |
|---|
|
 |
| EST |
nodata |
|
| GeneChip |
|
brain:7.32 | |
blood:6.74 | |
connective:8.82 | |
reproductive:6.33 | |
muscular:7.10 | |
alimentary:6.57 | |
liver:6.52 | |
lung:6.50 | |
urinary:6.37 | |
endo/exo-crine:6.98 | |
|
| CAGE |
|
brain:0.05 | |
blood:0.14 | |
connective:1.71 | |
reproductive:1.10 | |
muscular:0.23 | |
alimentary:0.06 | |
liver:0.00 | |
lung:0.00 | |
urinary:0.08 | |
endo/exo-crine:0.31 | |
|
| RNA-seq |
|
brain:2.59 | |
blood:1.91 | |
connective:6.78 | |
reproductive:1.84 | |
muscular:1.10 | |
alimentary:1.45 | |
liver:0.13 | |
lung:1.08 | |
urinary:0.63 | |
endo/exo-crine:3.41 | |
|
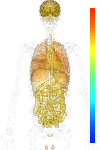
| Synonym | PHD1, EGL nine (C.elegans) homolog 2, DKFZp434E026, HIFPH1, HIF-PH1, EGLN2, Prolyl hydroxylase domain-containing protein 1, Estrogen-induced tag 6, HPH-3, egl nine homolog 2 (C. elegans), HIF-prolyl hydroxylase 1, Hypoxia-inducible factor prolyl hydroxylase 1, EIT6, Egl nine homolog 2 |
|---|
| Refseq ID | NM_053046 |
|---|
| GeneID | 112398 |
|---|
| Unigene ID | Hs.515417 |
|---|
| Probe set ID | 220956_s_at |
|---|
|
 |
| EST |
|
brain:191.60 | |
blood:339.00 | |
connective:122.10 | |
reproductive:281.80 | |
muscular:101.50 | |
alimentary:251.50 | |
liver:63.30 | |
lung:244.40 | |
urinary:124.80 | |
endo/exo-crine:190.30 | |
|
| GeneChip |
|
brain:7.84 | |
blood:8.12 | |
connective:7.93 | |
reproductive:7.95 | |
muscular:7.50 | |
alimentary:7.78 | |
liver:7.89 | |
lung:8.61 | |
urinary:7.66 | |
endo/exo-crine:7.89 | |
|
| CAGE |
|
brain:3.34 | |
blood:3.42 | |
connective:3.19 | |
reproductive:3.79 | |
muscular:3.21 | |
alimentary:3.33 | |
liver:3.07 | |
lung:3.86 | |
urinary:3.13 | |
endo/exo-crine:3.28 | |
|
| RNA-seq |
|
brain:0.00 | |
blood:0.00 | |
connective:0.04 | |
reproductive:1.72 | |
muscular:0.00 | |
alimentary:0.06 | |
liver:0.00 | |
lung:0.00 | |
urinary:0.00 | |
endo/exo-crine:0.14 | |
|
| Synonym | PHD1, EGL nine (C.elegans) homolog 2, DKFZp434E026, HIFPH1, HIF-PH1, EGLN2, Prolyl hydroxylase domain-containing protein 1, Estrogen-induced tag 6, HPH-3, egl nine homolog 2 (C. elegans), HIF-prolyl hydroxylase 1, Hypoxia-inducible factor prolyl hydroxylase 1, EIT6, Egl nine homolog 2 |
|---|
| Refseq ID | NM_080732 |
|---|
| GeneID | 112398 |
|---|
| Unigene ID | Hs.515417 |
|---|
| Probe set ID | 220956_s_at |
|---|
|
 |
| EST |
|
brain:191.60 | |
blood:339.00 | |
connective:122.10 | |
reproductive:281.80 | |
muscular:101.50 | |
alimentary:251.50 | |
liver:63.30 | |
lung:244.40 | |
urinary:124.80 | |
endo/exo-crine:190.30 | |
|
| GeneChip |
|
brain:7.84 | |
blood:8.12 | |
connective:7.93 | |
reproductive:7.95 | |
muscular:7.50 | |
alimentary:7.78 | |
liver:7.89 | |
lung:8.61 | |
urinary:7.66 | |
endo/exo-crine:7.89 | |
|
| CAGE |
|
brain:3.34 | |
blood:3.42 | |
connective:3.19 | |
reproductive:3.79 | |
muscular:3.21 | |
alimentary:3.33 | |
liver:3.07 | |
lung:3.86 | |
urinary:3.13 | |
endo/exo-crine:3.28 | |
|
| RNA-seq |
|
brain:3.01 | |
blood:4.72 | |
connective:4.06 | |
reproductive:4.70 | |
muscular:3.03 | |
alimentary:4.15 | |
liver:3.45 | |
lung:4.81 | |
urinary:4.02 | |
endo/exo-crine:4.25 | |
|
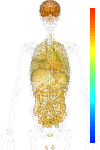
| Synonym | Glutamate receptor, ionotropic kainate 5 precursor, glutamate receptor, ionotropic, kainate 5, GRIK5, GRIK2, Glutamate receptor KA-2, KA2, EAA2, Excitatory amino acid receptor 2 |
|---|
| Refseq ID | NM_002088 |
|---|
| GeneID | 2901 |
|---|
| Unigene ID | Hs.367799 |
|---|
| Probe set ID | 217509_x_at |
|---|
|
 |
| EST |
|
brain:27.00 | |
blood:- | |
connective:6.40 | |
reproductive:10.90 | |
muscular:- | |
alimentary:- | |
liver:- | |
lung:- | |
urinary:- | |
endo/exo-crine:15.50 | |
|
| GeneChip |
|
brain:8.52 | |
blood:7.82 | |
connective:7.71 | |
reproductive:7.68 | |
muscular:8.09 | |
alimentary:7.92 | |
liver:8.12 | |
lung:7.76 | |
urinary:8.17 | |
endo/exo-crine:7.89 | |
|
| CAGE |
|
brain:0.13 | |
blood:0.00 | |
connective:0.00 | |
reproductive:0.00 | |
muscular:0.00 | |
alimentary:0.00 | |
liver:0.00 | |
lung:0.00 | |
urinary:0.00 | |
endo/exo-crine:0.00 | |
|
| RNA-seq |
|
brain:2.81 | |
blood:1.00 | |
connective:0.31 | |
reproductive:2.15 | |
muscular:0.37 | |
alimentary:1.91 | |
liver:0.02 | |
lung:0.39 | |
urinary:1.57 | |
endo/exo-crine:1.45 | |
|
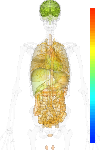
| Synonym | rRNA 2'-O-methyltransferase fibrillarin, 34 kDa nucleolar scleroderma antigen, FBL, RNU3IP1, fibrillarin, FLRN, FIB1, FIB |
|---|
| Refseq ID | NM_001436 |
|---|
| GeneID | 2091 |
|---|
| Unigene ID | Hs.299002 |
|---|
| Probe set ID | 211623_s_at |
|---|
|
 |
| EST |
|
brain:33.80 | |
blood:135.60 | |
connective:99.60 | |
reproductive:79.30 | |
muscular:132.70 | |
alimentary:115.70 | |
liver:42.20 | |
lung:88.00 | |
urinary:28.80 | |
endo/exo-crine:62.10 | |
|
| GeneChip |
|
brain:6.99 | |
blood:8.37 | |
connective:8.37 | |
reproductive:8.45 | |
muscular:7.46 | |
alimentary:8.22 | |
liver:7.00 | |
lung:7.88 | |
urinary:7.49 | |
endo/exo-crine:8.44 | |
|
| CAGE |
|
brain:3.63 | |
blood:4.95 | |
connective:4.47 | |
reproductive:4.70 | |
muscular:4.56 | |
alimentary:4.81 | |
liver:4.14 | |
lung:4.64 | |
urinary:4.47 | |
endo/exo-crine:4.35 | |
|
| RNA-seq |
|
brain:4.46 | |
blood:6.50 | |
connective:6.75 | |
reproductive:6.87 | |
muscular:4.98 | |
alimentary:6.53 | |
liver:4.85 | |
lung:6.41 | |
urinary:5.37 | |
endo/exo-crine:6.39 | |
|