| Synonym | Slit-2, SLIT2, Slit homolog 2 protein precursor, slit (Drosophila) homolog 2, FLJ14420, SLIL3, slit homolog 2 (Drosophila) |
|---|
| Refseq ID | NM_004787 |
|---|
| GeneID | 9353 |
|---|
| Unigene ID | - |
|---|
| Probe set ID | 209897_s_at |
|---|
|
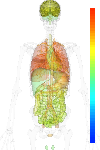 |
| EST |
nodata |
|
| GeneChip |
|
brain:7.03 | |
blood:7.44 | |
connective:7.37 | |
reproductive:7.68 | |
muscular:6.91 | |
alimentary:7.34 | |
liver:5.54 | |
lung:9.86 | |
urinary:7.71 | |
endo/exo-crine:7.72 | |
|
| CAGE |
|
brain:0.85 | |
blood:0.88 | |
connective:0.90 | |
reproductive:1.35 | |
muscular:0.60 | |
alimentary:1.10 | |
liver:0.14 | |
lung:2.57 | |
urinary:1.50 | |
endo/exo-crine:0.72 | |
|
| RNA-seq |
|
brain:1.75 | |
blood:2.42 | |
connective:1.60 | |
reproductive:2.41 | |
muscular:1.04 | |
alimentary:2.95 | |
liver:0.26 | |
lung:4.32 | |
urinary:1.92 | |
endo/exo-crine:2.11 | |
|
| Synonym | HUMORFA01, IQ motif containing GTPase activating protein 1, SAR1, IQGAP1, Ras GTPase-activating-like protein IQGAP1, p195, KIAA0051 |
|---|
| Refseq ID | NM_003870 |
|---|
| GeneID | 8826 |
|---|
| Unigene ID | Hs.430551 |
|---|
| Probe set ID | 200791_s_at |
|---|
|
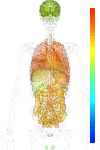 |
| EST |
|
brain:66.70 | |
blood:208.60 | |
connective:285.90 | |
reproductive:161.40 | |
muscular:70.30 | |
alimentary:191.20 | |
liver:242.80 | |
lung:210.20 | |
urinary:105.60 | |
endo/exo-crine:225.20 | |
|
| GeneChip |
|
brain:6.91 | |
blood:8.75 | |
connective:8.93 | |
reproductive:8.75 | |
muscular:7.68 | |
alimentary:8.42 | |
liver:6.65 | |
lung:9.12 | |
urinary:7.67 | |
endo/exo-crine:8.52 | |
|
| CAGE |
|
brain:4.17 | |
blood:5.41 | |
connective:5.61 | |
reproductive:5.07 | |
muscular:4.88 | |
alimentary:5.18 | |
liver:3.93 | |
lung:5.50 | |
urinary:5.25 | |
endo/exo-crine:5.00 | |
|
| RNA-seq |
|
brain:3.62 | |
blood:5.44 | |
connective:5.61 | |
reproductive:5.41 | |
muscular:3.65 | |
alimentary:5.58 | |
liver:2.58 | |
lung:5.76 | |
urinary:5.10 | |
endo/exo-crine:5.23 | |
|
| Synonym | IQGAP2, Ras GTPase-activating-like protein IQGAP2, IQ motif containing GTPase activating protein 2 |
|---|
| Refseq ID | NM_006633 |
|---|
| GeneID | 10788 |
|---|
| Unigene ID | Hs.291030 |
|---|
| Probe set ID | 203474_at |
|---|
|
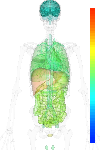 |
| EST |
|
brain:22.80 | |
blood:39.10 | |
connective:35.30 | |
reproductive:138.20 | |
muscular:7.80 | |
alimentary:90.50 | |
liver:105.60 | |
lung:44.00 | |
urinary:115.20 | |
endo/exo-crine:58.20 | |
|
| GeneChip |
|
brain:3.81 | |
blood:5.52 | |
connective:5.00 | |
reproductive:5.97 | |
muscular:4.31 | |
alimentary:6.05 | |
liver:8.93 | |
lung:5.63 | |
urinary:6.91 | |
endo/exo-crine:6.22 | |
|
| CAGE |
|
brain:1.01 | |
blood:3.16 | |
connective:3.38 | |
reproductive:3.28 | |
muscular:2.06 | |
alimentary:3.63 | |
liver:4.96 | |
lung:3.17 | |
urinary:3.04 | |
endo/exo-crine:2.91 | |
|
| RNA-seq |
|
brain:0.27 | |
blood:2.77 | |
connective:1.78 | |
reproductive:3.12 | |
muscular:0.88 | |
alimentary:2.41 | |
liver:5.17 | |
lung:1.77 | |
urinary:3.77 | |
endo/exo-crine:2.65 | |
|
| Synonym | SCIP1, CDC42SE1, SPEC1, CDC42 small effector 1 |
|---|
| Refseq ID | NM_001038707 |
|---|
| GeneID | 56882 |
|---|
| Unigene ID | Hs.22065 |
|---|
| Probe set ID | 218157_x_at |
|---|
|
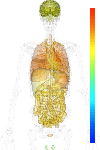 |
| EST |
|
brain:40.50 | |
blood:122.60 | |
connective:106.00 | |
reproductive:65.70 | |
muscular:31.20 | |
alimentary:60.40 | |
liver:42.20 | |
lung:117.30 | |
urinary:28.80 | |
endo/exo-crine:89.30 | |
|
| GeneChip |
|
brain:7.41 | |
blood:8.52 | |
connective:7.90 | |
reproductive:7.78 | |
muscular:7.19 | |
alimentary:8.01 | |
liver:7.38 | |
lung:8.82 | |
urinary:7.64 | |
endo/exo-crine:8.08 | |
|
| CAGE |
|
brain:4.06 | |
blood:4.85 | |
connective:4.42 | |
reproductive:4.05 | |
muscular:4.10 | |
alimentary:4.16 | |
liver:3.60 | |
lung:4.59 | |
urinary:4.22 | |
endo/exo-crine:4.22 | |
|
| RNA-seq |
|
brain:0.68 | |
blood:1.74 | |
connective:0.65 | |
reproductive:2.03 | |
muscular:1.20 | |
alimentary:1.52 | |
liver:0.58 | |
lung:1.99 | |
urinary:2.76 | |
endo/exo-crine:1.92 | |
|
| Synonym | SCIP1, CDC42SE1, SPEC1, CDC42 small effector 1 |
|---|
| Refseq ID | NM_020239 |
|---|
| GeneID | 56882 |
|---|
| Unigene ID | Hs.22065 |
|---|
| Probe set ID | 218157_x_at |
|---|
|
 |
| EST |
|
brain:40.50 | |
blood:122.60 | |
connective:106.00 | |
reproductive:65.70 | |
muscular:31.20 | |
alimentary:60.40 | |
liver:42.20 | |
lung:117.30 | |
urinary:28.80 | |
endo/exo-crine:89.30 | |
|
| GeneChip |
|
brain:7.41 | |
blood:8.52 | |
connective:7.90 | |
reproductive:7.78 | |
muscular:7.19 | |
alimentary:8.01 | |
liver:7.38 | |
lung:8.82 | |
urinary:7.64 | |
endo/exo-crine:8.08 | |
|
| CAGE |
|
brain:4.06 | |
blood:4.85 | |
connective:4.42 | |
reproductive:4.05 | |
muscular:4.10 | |
alimentary:4.16 | |
liver:3.60 | |
lung:4.59 | |
urinary:4.22 | |
endo/exo-crine:4.22 | |
|
| RNA-seq |
|
brain:3.53 | |
blood:5.90 | |
connective:4.39 | |
reproductive:4.15 | |
muscular:3.02 | |
alimentary:3.70 | |
liver:3.24 | |
lung:5.56 | |
urinary:4.17 | |
endo/exo-crine:4.23 | |
|
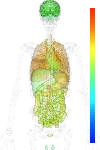
| Synonym | HUMORFA01, IQ motif containing GTPase activating protein 1, SAR1, IQGAP1, Ras GTPase-activating-like protein IQGAP1, p195, KIAA0051 |
|---|
| Refseq ID | NM_003870 |
|---|
| GeneID | 8826 |
|---|
| Unigene ID | Hs.430551 |
|---|
| Probe set ID | 213446_s_at |
|---|
|
 |
| EST |
|
brain:66.70 | |
blood:208.60 | |
connective:285.90 | |
reproductive:161.40 | |
muscular:70.30 | |
alimentary:191.20 | |
liver:242.80 | |
lung:210.20 | |
urinary:105.60 | |
endo/exo-crine:225.20 | |
|
| GeneChip |
|
brain:5.63 | |
blood:7.53 | |
connective:6.92 | |
reproductive:7.38 | |
muscular:6.30 | |
alimentary:7.47 | |
liver:5.72 | |
lung:8.40 | |
urinary:6.72 | |
endo/exo-crine:7.30 | |
|
| CAGE |
|
brain:4.17 | |
blood:5.41 | |
connective:5.61 | |
reproductive:5.07 | |
muscular:4.88 | |
alimentary:5.18 | |
liver:3.93 | |
lung:5.50 | |
urinary:5.25 | |
endo/exo-crine:5.00 | |
|
| RNA-seq |
|
brain:3.62 | |
blood:5.44 | |
connective:5.61 | |
reproductive:5.41 | |
muscular:3.65 | |
alimentary:5.58 | |
liver:2.58 | |
lung:5.76 | |
urinary:5.10 | |
endo/exo-crine:5.23 | |
|
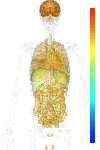
| Synonym | ADP-ribosylation factor-like 2, ARL2, ADP-ribosylation factor-like protein 2, ARFL2 |
|---|
| Refseq ID | NM_001667 |
|---|
| GeneID | 402 |
|---|
| Unigene ID | Hs.502836 |
|---|
| Probe set ID | 202564_x_at |
|---|
|
 |
| EST |
|
brain:34.60 | |
blood:18.30 | |
connective:41.80 | |
reproductive:28.70 | |
muscular:62.50 | |
alimentary:166.00 | |
liver:- | |
lung:39.10 | |
urinary:28.80 | |
endo/exo-crine:23.30 | |
|
| GeneChip |
|
brain:8.38 | |
blood:7.94 | |
connective:8.27 | |
reproductive:7.78 | |
muscular:8.16 | |
alimentary:7.82 | |
liver:6.79 | |
lung:7.90 | |
urinary:8.37 | |
endo/exo-crine:7.78 | |
|
| CAGE |
|
brain:3.35 | |
blood:2.37 | |
connective:3.03 | |
reproductive:3.08 | |
muscular:3.11 | |
alimentary:2.69 | |
liver:1.44 | |
lung:2.63 | |
urinary:3.19 | |
endo/exo-crine:2.40 | |
|
| RNA-seq |
|
brain:5.45 | |
blood:5.46 | |
connective:6.15 | |
reproductive:5.98 | |
muscular:5.39 | |
alimentary:5.71 | |
liver:2.72 | |
lung:5.11 | |
urinary:5.43 | |
endo/exo-crine:5.86 | |
|
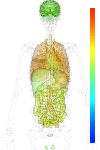
| Synonym | HUMORFA01, IQ motif containing GTPase activating protein 1, SAR1, IQGAP1, Ras GTPase-activating-like protein IQGAP1, p195, KIAA0051 |
|---|
| Refseq ID | NM_003870 |
|---|
| GeneID | 8826 |
|---|
| Unigene ID | Hs.430551 |
|---|
| Probe set ID | 210840_s_at |
|---|
|
 |
| EST |
|
brain:66.70 | |
blood:208.60 | |
connective:285.90 | |
reproductive:161.40 | |
muscular:70.30 | |
alimentary:191.20 | |
liver:242.80 | |
lung:210.20 | |
urinary:105.60 | |
endo/exo-crine:225.20 | |
|
| GeneChip |
|
brain:6.17 | |
blood:7.89 | |
connective:7.80 | |
reproductive:7.94 | |
muscular:6.61 | |
alimentary:7.60 | |
liver:6.12 | |
lung:8.31 | |
urinary:6.73 | |
endo/exo-crine:7.58 | |
|
| CAGE |
|
brain:4.17 | |
blood:5.41 | |
connective:5.61 | |
reproductive:5.07 | |
muscular:4.88 | |
alimentary:5.18 | |
liver:3.93 | |
lung:5.50 | |
urinary:5.25 | |
endo/exo-crine:5.00 | |
|
| RNA-seq |
|
brain:3.62 | |
blood:5.44 | |
connective:5.61 | |
reproductive:5.41 | |
muscular:3.65 | |
alimentary:5.58 | |
liver:2.58 | |
lung:5.76 | |
urinary:5.10 | |
endo/exo-crine:5.23 | |
|
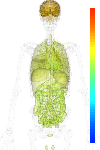
| Synonym | tyrosine kinase, non-receptor, 2, p21cdc42Hs, ACK1, TNK2, ACK-1, FLJ45547, FLJ44758, Tyrosine kinase non-receptor protein 2, ACK, Activated CDC42 kinase 1 |
|---|
| Refseq ID | NM_001010938 |
|---|
| GeneID | 10188 |
|---|
| Unigene ID | Hs.518513 |
|---|
| Probe set ID | 203839_s_at |
|---|
|
 |
| EST |
|
brain:79.40 | |
blood:120.00 | |
connective:45.00 | |
reproductive:78.00 | |
muscular:46.80 | |
alimentary:90.50 | |
liver:- | |
lung:34.20 | |
urinary:28.80 | |
endo/exo-crine:69.90 | |
|
| GeneChip |
|
brain:7.84 | |
blood:7.50 | |
connective:7.16 | |
reproductive:6.81 | |
muscular:6.99 | |
alimentary:7.16 | |
liver:7.31 | |
lung:7.31 | |
urinary:6.69 | |
endo/exo-crine:7.28 | |
|
| CAGE |
|
brain:2.45 | |
blood:1.18 | |
connective:1.01 | |
reproductive:1.11 | |
muscular:1.09 | |
alimentary:1.41 | |
liver:0.84 | |
lung:1.12 | |
urinary:0.71 | |
endo/exo-crine:0.77 | |
|
| RNA-seq |
|
brain:1.86 | |
blood:1.14 | |
connective:0.11 | |
reproductive:1.06 | |
muscular:1.19 | |
alimentary:0.27 | |
liver:0.38 | |
lung:0.67 | |
urinary:0.00 | |
endo/exo-crine:0.88 | |
|
| Synonym | tyrosine kinase, non-receptor, 2, p21cdc42Hs, ACK1, TNK2, ACK-1, FLJ45547, FLJ44758, Tyrosine kinase non-receptor protein 2, ACK, Activated CDC42 kinase 1 |
|---|
| Refseq ID | NM_005781 |
|---|
| GeneID | 10188 |
|---|
| Unigene ID | Hs.518513 |
|---|
| Probe set ID | 203839_s_at |
|---|
|
 |
| EST |
|
brain:79.40 | |
blood:120.00 | |
connective:45.00 | |
reproductive:78.00 | |
muscular:46.80 | |
alimentary:90.50 | |
liver:- | |
lung:34.20 | |
urinary:28.80 | |
endo/exo-crine:69.90 | |
|
| GeneChip |
|
brain:7.84 | |
blood:7.50 | |
connective:7.16 | |
reproductive:6.81 | |
muscular:6.99 | |
alimentary:7.16 | |
liver:7.31 | |
lung:7.31 | |
urinary:6.69 | |
endo/exo-crine:7.28 | |
|
| CAGE |
|
brain:2.45 | |
blood:1.18 | |
connective:1.01 | |
reproductive:1.11 | |
muscular:1.09 | |
alimentary:1.41 | |
liver:0.84 | |
lung:1.12 | |
urinary:0.71 | |
endo/exo-crine:0.77 | |
|
| RNA-seq |
|
brain:3.16 | |
blood:3.54 | |
connective:2.30 | |
reproductive:2.43 | |
muscular:1.39 | |
alimentary:2.44 | |
liver:1.43 | |
lung:2.36 | |
urinary:1.14 | |
endo/exo-crine:2.45 | |
|