| Synonym | FLJ20300, Glycine N-choloyltransferase, Long-chain fatty-acyl-CoA hydrolase, BAT, MGC104432, BACAT, BAAT, Bile acid CoA:amino acid N-acyltransferase |
|---|
| Refseq ID | NM_001127610 |
|---|
| GeneID | 570 |
|---|
| Unigene ID | - |
|---|
| Probe set ID | 206913_at |
|---|
|
 |
| EST |
nodata |
|
| GeneChip |
|
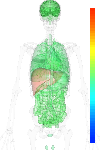
brain:5.33 | |
blood:5.07 | |
connective:5.19 | |
reproductive:4.93 | |
muscular:5.43 | |
alimentary:5.22 | |
liver:10.33 | |
lung:4.94 | |
urinary:5.19 | |
endo/exo-crine:4.97 | |
|
| CAGE |
|
brain:0.10 | |
blood:0.04 | |
connective:0.00 | |
reproductive:0.00 | |
muscular:0.11 | |
alimentary:0.00 | |
liver:5.65 | |
lung:0.14 | |
urinary:0.07 | |
endo/exo-crine:0.10 | |
|
| RNA-seq |
|
brain:0.00 | |
blood:0.04 | |
connective:0.01 | |
reproductive:0.03 | |
muscular:0.03 | |
alimentary:0.00 | |
liver:4.91 | |
lung:0.00 | |
urinary:0.04 | |
endo/exo-crine:0.02 | |
|
| Synonym | FLJ20300, Glycine N-choloyltransferase, Long-chain fatty-acyl-CoA hydrolase, BAT, MGC104432, BACAT, BAAT, Bile acid CoA:amino acid N-acyltransferase |
|---|
| Refseq ID | NM_001127610 |
|---|
| GeneID | 570 |
|---|
| Unigene ID | Hs.284712 |
|---|
| Probe set ID | 206913_at |
|---|
|
 |
| EST |
|
brain:- | |
blood:- | |
connective:- | |
reproductive:- | |
muscular:15.60 | |
alimentary:- | |
liver:232.20 | |
lung:9.80 | |
urinary:9.60 | |
endo/exo-crine:- | |
|
| GeneChip |
|
brain:5.33 | |
blood:5.07 | |
connective:5.19 | |
reproductive:4.93 | |
muscular:5.43 | |
alimentary:5.22 | |
liver:10.33 | |
lung:4.94 | |
urinary:5.19 | |
endo/exo-crine:4.97 | |
|
| CAGE |
|
brain:0.10 | |
blood:0.04 | |
connective:0.00 | |
reproductive:0.00 | |
muscular:0.11 | |
alimentary:0.00 | |
liver:5.65 | |
lung:0.14 | |
urinary:0.07 | |
endo/exo-crine:0.10 | |
|
| RNA-seq |
|
brain:0.00 | |
blood:0.04 | |
connective:0.01 | |
reproductive:0.03 | |
muscular:0.03 | |
alimentary:0.00 | |
liver:4.91 | |
lung:0.00 | |
urinary:0.04 | |
endo/exo-crine:0.02 | |
|
| Synonym | FLJ20300, Glycine N-choloyltransferase, Long-chain fatty-acyl-CoA hydrolase, BAT, MGC104432, BACAT, BAAT, Bile acid CoA:amino acid N-acyltransferase |
|---|
| Refseq ID | NM_001701 |
|---|
| GeneID | 570 |
|---|
| Unigene ID | - |
|---|
| Probe set ID | 206913_at |
|---|
|
 |
| EST |
nodata |
|
| GeneChip |
|
brain:5.33 | |
blood:5.07 | |
connective:5.19 | |
reproductive:4.93 | |
muscular:5.43 | |
alimentary:5.22 | |
liver:10.33 | |
lung:4.94 | |
urinary:5.19 | |
endo/exo-crine:4.97 | |
|
| CAGE |
|
brain:0.10 | |
blood:0.04 | |
connective:0.00 | |
reproductive:0.00 | |
muscular:0.11 | |
alimentary:0.00 | |
liver:5.65 | |
lung:0.14 | |
urinary:0.07 | |
endo/exo-crine:0.10 | |
|
| RNA-seq |
|
brain:0.69 | |
blood:0.05 | |
connective:0.00 | |
reproductive:0.00 | |
muscular:0.01 | |
alimentary:0.00 | |
liver:6.15 | |
lung:0.00 | |
urinary:0.00 | |
endo/exo-crine:0.01 | |
|
| Synonym | FLJ20300, Glycine N-choloyltransferase, Long-chain fatty-acyl-CoA hydrolase, BAT, MGC104432, BACAT, BAAT, Bile acid CoA:amino acid N-acyltransferase |
|---|
| Refseq ID | NM_001701 |
|---|
| GeneID | 570 |
|---|
| Unigene ID | Hs.284712 |
|---|
| Probe set ID | 206913_at |
|---|
|
 |
| EST |
|
brain:- | |
blood:- | |
connective:- | |
reproductive:- | |
muscular:15.60 | |
alimentary:- | |
liver:232.20 | |
lung:9.80 | |
urinary:9.60 | |
endo/exo-crine:- | |
|
| GeneChip |
|
brain:5.33 | |
blood:5.07 | |
connective:5.19 | |
reproductive:4.93 | |
muscular:5.43 | |
alimentary:5.22 | |
liver:10.33 | |
lung:4.94 | |
urinary:5.19 | |
endo/exo-crine:4.97 | |
|
| CAGE |
|
brain:0.10 | |
blood:0.04 | |
connective:0.00 | |
reproductive:0.00 | |
muscular:0.11 | |
alimentary:0.00 | |
liver:5.65 | |
lung:0.14 | |
urinary:0.07 | |
endo/exo-crine:0.10 | |
|
| RNA-seq |
|
brain:0.69 | |
blood:0.05 | |
connective:0.00 | |
reproductive:0.00 | |
muscular:0.01 | |
alimentary:0.00 | |
liver:6.15 | |
lung:0.00 | |
urinary:0.00 | |
endo/exo-crine:0.01 | |
|
| Synonym | Palmitoyl-protein hydrolase 1, Palmitoyl-protein thioesterase 1 precursor, PPT-1, INCL, CLN1, PPT1 |
|---|
| Refseq ID | NM_000310 |
|---|
| GeneID | 5538 |
|---|
| Unigene ID | Hs.387300 |
|---|
| Probe set ID | 200975_at |
|---|
|
 |
| EST |
|
brain:46.40 | |
blood:- | |
connective:3.20 | |
reproductive:8.20 | |
muscular:7.80 | |
alimentary:- | |
liver:- | |
lung:9.80 | |
urinary:- | |
endo/exo-crine:3.90 | |
|
| GeneChip |
|
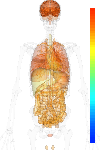
brain:9.30 | |
blood:8.74 | |
connective:8.47 | |
reproductive:8.98 | |
muscular:7.73 | |
alimentary:8.28 | |
liver:7.40 | |
lung:9.52 | |
urinary:8.65 | |
endo/exo-crine:8.79 | |
|
| CAGE |
|
brain:4.67 | |
blood:4.50 | |
connective:4.70 | |
reproductive:4.12 | |
muscular:3.74 | |
alimentary:3.66 | |
liver:3.78 | |
lung:4.90 | |
urinary:4.14 | |
endo/exo-crine:4.13 | |
|
| RNA-seq |
|
brain:5.76 | |
blood:5.12 | |
connective:4.91 | |
reproductive:5.48 | |
muscular:3.33 | |
alimentary:5.03 | |
liver:3.56 | |
lung:5.67 | |
urinary:5.24 | |
endo/exo-crine:5.22 | |
|
| Synonym | acyl-CoA thioesterase 2, ZAP128, PTE2, CTE-Ia, ACOT2, Peroxisomal acyl-coenzyme A thioester hydrolase 2a, Acyl-coenzyme A thioesterase 2, Acyl-CoA thioesterase 2, Mte1, Peroxisomal long-chain acyl-coA thioesterase 2, PTE2A |
|---|
| Refseq ID | NM_006821 |
|---|
| GeneID | 10965 |
|---|
| Unigene ID | Hs.446685 |
|---|
| Probe set ID | 202982_s_at |
|---|
|
 |
| EST |
|
brain:43.90 | |
blood:54.80 | |
connective:93.20 | |
reproductive:68.40 | |
muscular:117.10 | |
alimentary:60.40 | |
liver:10.60 | |
lung:73.30 | |
urinary:76.80 | |
endo/exo-crine:11.60 | |
|
| GeneChip |
|
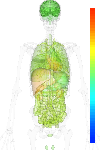
brain:5.83 | |
blood:6.19 | |
connective:8.03 | |
reproductive:6.37 | |
muscular:7.81 | |
alimentary:6.42 | |
liver:8.37 | |
lung:6.51 | |
urinary:7.85 | |
endo/exo-crine:6.62 | |
|
| CAGE |
|
brain:2.15 | |
blood:2.30 | |
connective:3.70 | |
reproductive:2.55 | |
muscular:3.42 | |
alimentary:3.20 | |
liver:4.41 | |
lung:2.44 | |
urinary:3.10 | |
endo/exo-crine:2.14 | |
|
| RNA-seq |
|
brain:2.69 | |
blood:2.27 | |
connective:4.06 | |
reproductive:2.87 | |
muscular:4.09 | |
alimentary:3.00 | |
liver:5.09 | |
lung:2.69 | |
urinary:4.29 | |
endo/exo-crine:4.14 | |
|
| Synonym | ACH2, ACOT1, LACH2, CTE-1, acyl-CoA thioesterase 1 |
|---|
| Refseq ID | NM_001037161 |
|---|
| GeneID | 641371 |
|---|
| Unigene ID | Hs.568046 |
|---|
| Probe set ID | 202982_s_at |
|---|
|
 |
| EST |
|
brain:14.40 | |
blood:2.60 | |
connective:25.70 | |
reproductive:52.00 | |
muscular:46.80 | |
alimentary:10.10 | |
liver:10.60 | |
lung:24.40 | |
urinary:9.60 | |
endo/exo-crine:19.40 | |
|
| GeneChip |
|
brain:5.83 | |
blood:6.19 | |
connective:8.03 | |
reproductive:6.37 | |
muscular:7.81 | |
alimentary:6.42 | |
liver:8.37 | |
lung:6.51 | |
urinary:7.85 | |
endo/exo-crine:6.62 | |
|
| CAGE |
|
brain:0.34 | |
blood:0.18 | |
connective:0.78 | |
reproductive:0.45 | |
muscular:0.71 | |
alimentary:0.70 | |
liver:1.69 | |
lung:0.74 | |
urinary:0.75 | |
endo/exo-crine:0.27 | |
|
| RNA-seq |
|
brain:2.15 | |
blood:1.89 | |
connective:3.35 | |
reproductive:2.65 | |
muscular:3.12 | |
alimentary:2.47 | |
liver:4.38 | |
lung:2.06 | |
urinary:3.79 | |
endo/exo-crine:3.52 | |
|
| Synonym | DAPAT, Dihydroxyacetone phosphate acyltransferase, DHAPAT, Glycerone-phosphate O-acyltransferase, DAP-AT, DHAP-AT, GNPAT, glyceronephosphate O-acyltransferase, Acyl-CoA:dihydroxyacetonephosphateacyltransferase |
|---|
| Refseq ID | NM_014236 |
|---|
| GeneID | 8443 |
|---|
| Unigene ID | Hs.498028 |
|---|
| Probe set ID | 201956_s_at |
|---|
|
 |
| EST |
|
brain:109.80 | |
blood:120.00 | |
connective:93.20 | |
reproductive:128.60 | |
muscular:132.70 | |
alimentary:100.60 | |
liver:31.70 | |
lung:88.00 | |
urinary:153.60 | |
endo/exo-crine:93.20 | |
|
| GeneChip |
|
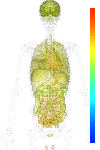
brain:7.22 | |
blood:7.45 | |
connective:7.58 | |
reproductive:7.74 | |
muscular:8.29 | |
alimentary:7.52 | |
liver:7.43 | |
lung:7.37 | |
urinary:7.42 | |
endo/exo-crine:7.47 | |
|
| CAGE |
|
brain:3.81 | |
blood:3.68 | |
connective:3.68 | |
reproductive:3.55 | |
muscular:4.29 | |
alimentary:3.81 | |
liver:3.33 | |
lung:3.83 | |
urinary:3.49 | |
endo/exo-crine:3.86 | |
|
| RNA-seq |
|
brain:3.31 | |
blood:4.24 | |
connective:4.09 | |
reproductive:4.58 | |
muscular:5.01 | |
alimentary:4.59 | |
liver:3.65 | |
lung:3.48 | |
urinary:4.29 | |
endo/exo-crine:4.24 | |
|
| Synonym | hBACH, CTE-IIa, LACH, acyl-CoA thioesterase 7, LACH1, Brain acyl-CoA hydrolase, ACOT7, ACH1, BACH, MGC1126, Long chain acyl-CoA thioester hydrolase, Cytosolic acyl coenzyme A thioester hydrolase, CTE-II, Acyl-CoA thioesterase 7 |
|---|
| Refseq ID | NM_007274 |
|---|
| GeneID | 11332 |
|---|
| Unigene ID | Hs.126137 |
|---|
| Probe set ID | 208002_s_at |
|---|
|
 |
| EST |
|
brain:75.10 | |
blood:20.90 | |
connective:41.80 | |
reproductive:57.50 | |
muscular:23.40 | |
alimentary:70.40 | |
liver:21.10 | |
lung:19.60 | |
urinary:38.40 | |
endo/exo-crine:27.20 | |
|
| GeneChip |
|
brain:8.07 | |
blood:6.16 | |
connective:6.02 | |
reproductive:5.67 | |
muscular:5.16 | |
alimentary:6.25 | |
liver:5.72 | |
lung:6.35 | |
urinary:7.37 | |
endo/exo-crine:5.79 | |
|
| CAGE |
|
brain:3.89 | |
blood:2.96 | |
connective:2.46 | |
reproductive:2.96 | |
muscular:2.48 | |
alimentary:3.38 | |
liver:2.77 | |
lung:2.96 | |
urinary:3.19 | |
endo/exo-crine:2.57 | |
|
| RNA-seq |
|
brain:0.92 | |
blood:1.32 | |
connective:0.66 | |
reproductive:0.90 | |
muscular:0.58 | |
alimentary:0.77 | |
liver:0.73 | |
lung:1.19 | |
urinary:0.31 | |
endo/exo-crine:0.43 | |
|
| Synonym | hBACH, CTE-IIa, LACH, acyl-CoA thioesterase 7, LACH1, Brain acyl-CoA hydrolase, ACOT7, ACH1, BACH, MGC1126, Long chain acyl-CoA thioester hydrolase, Cytosolic acyl coenzyme A thioester hydrolase, CTE-II, Acyl-CoA thioesterase 7 |
|---|
| Refseq ID | NM_181864 |
|---|
| GeneID | 11332 |
|---|
| Unigene ID | Hs.126137 |
|---|
| Probe set ID | 208002_s_at |
|---|
|
 |
| EST |
|
brain:75.10 | |
blood:20.90 | |
connective:41.80 | |
reproductive:57.50 | |
muscular:23.40 | |
alimentary:70.40 | |
liver:21.10 | |
lung:19.60 | |
urinary:38.40 | |
endo/exo-crine:27.20 | |
|
| GeneChip |
|
brain:8.07 | |
blood:6.16 | |
connective:6.02 | |
reproductive:5.67 | |
muscular:5.16 | |
alimentary:6.25 | |
liver:5.72 | |
lung:6.35 | |
urinary:7.37 | |
endo/exo-crine:5.79 | |
|
| CAGE |
|
brain:3.89 | |
blood:2.96 | |
connective:2.46 | |
reproductive:2.96 | |
muscular:2.48 | |
alimentary:3.38 | |
liver:2.77 | |
lung:2.96 | |
urinary:3.19 | |
endo/exo-crine:2.57 | |
|
| RNA-seq |
|
brain:1.47 | |
blood:0.00 | |
connective:0.14 | |
reproductive:0.23 | |
muscular:0.02 | |
alimentary:0.99 | |
liver:0.00 | |
lung:0.48 | |
urinary:0.66 | |
endo/exo-crine:0.39 | |
|